More Information
Submitted: September 09, 2024 | Approved: September 12, 2024 | Published: September 13, 2024
How to cite this article: Ogunbiyi TE, Mustapha AM, Eturhobore EJ, Achas MJ, Sessi TA. Development of a Web-based Tomato Plant Disease Detection and Diagnosis System using Transfer Learning Techniques. Ann Civil Environ Eng. 2024; 8(1): 076-086. Available from: https://dx.doi.org/10.29328/journal.acee.1001071
DOI: 10.29328/journal.acee.1001071
Copyright License: © 2024 Ogunbiyi TE, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Plant disease detection; Tomato leaf; Transfer learning; Sustainable farming; CNN; DenseNet121; VGG16; VGG19; Sustainable agriculture
Development of a Web-based Tomato Plant Disease Detection and Diagnosis System using Transfer Learning Techniques
TE Ogunbiyi1* , AM Mustapha2
, AM Mustapha2 , EJ Eturhobore1, MJ Achas1 and TA Sessi3
, EJ Eturhobore1, MJ Achas1 and TA Sessi3
1Department of Computer Science and Information Technology, Bells University of Technology, Ota, Nigeria
2Department of Mathematical Sciences, Anchor University, Lagos, Nigeria
3Department of Agriculture and Agricultural Technology (AAT), Bells University of Technology, Ota, Nigeria
*Address for Correspondence: TE Ogunbiyi, Department of Computer Science and Information Technology, Bells University of Technology, Ota, Nigeria, Email: [email protected]
A significant obstacle to agricultural productivity that jeopardizes the availability of food is crop diseases and farmer livelihoods by reducing crop yields. Traditional visual assessment methods for disease diagnosis are effective but complex, often requiring expert observers. Recent advancements in deep learning indicate the potential for increasing accuracy and automating disease identification. Developing accessible diagnostic tools, such as web applications leveraging CNNs, can provide farmers with efficient and accurate disease identification, especially in regions with limited access to advanced diagnostic technologies. The main goal is to develop a productive system that can recognize tomato plant diseases. The model was trained on a collection of images of healthy and damaged tomato leaves from PlantVillage using transfer learning techniques. The images from the dataset were cleansed by resizing them from 256 × 256 to 224 × 224 to match the dimensions used in pre-trained models using min-max normalization. An evaluation of VGG16, VGG19, and DenseNet121 models based on performance accuracy and loss value for 7 categories of tomatoes guided the selection of the most effective model for practical application. VGG16 achieved 84.54% accuracy, VGG19 achieved 84.62%, and DenseNet121 achieved 98.28%, making DenseNet121 the chosen model due to its highest performance accuracy. The web application development based on the DenseNet121 architecture was integrated using the Django web framework, which is built on Python. This enables real-time disease diagnosis for uploaded images of tomato leaves. The proposed system allows early detection and diagnosis of tomato plant diseases, helping to mitigate crop losses. This supports sustainable farming practices and increases agricultural productivity.
Tomato is a species of the Solanaceae family and the second-highest widely grown crop in the United States of America, after potatoes. The plants often grow to a height of 1 - 3 meters (3 feet - 10 feet), with a weak stem that stretches over the ground and climbs over nearby plants [1]. Human beings eat tomatoes without restriction because they help the heart and other organs as a result of Lycopene, a naturally occurring antioxidant, present in them. Lycopene, which is largely found in cooked tomatoes, has been shown in some studies to improve the skin's protection against UV light and might help against prostate tumors [2]. Apart from the health benefits, tomatoes generate high income for the farmers and industrialists who process them [3]. Crops grown for food, inclusive of tomatoes, are susceptible to infections, which can be brought on by bacterial, fungal, or viral pathogens leading to plant diseases. Plant diseases represent an organic phenomenon that limits the development, proliferation, and development of plants [4]. Plant diseases and the microbes responsible for them pose a direct threat to the world's sustenance and prosperity [5] contradicting SDGs 1 and 2 (eradication of poverty and zero hunger). A major barrier to agricultural crop productivity is plant disease [6]. They threaten both the world's food supply and the livelihood of the farmers who grow the crops since they reduce plant productivity. According to research, illnesses cause an annual loss of almost 25% of the world's crop yield [7]. The economic burden of disease-related losses has led to the abandonment of entire plantations in locations like Panama [8]. The farmers experience substantial financial and economic losses as a result of these losses. In a similar vein, it can result in starvation, which in turn might cause death. Plant diseases can have disastrous socio-economic effects; examples include the Bengal famine pandemic of 1943 and the potato blight that struck Ireland in 1845–1846, according to published research.
Diseases of plants can cause physical alterations to their roots, leaves, stems, and other parts. Expert observers are capable of identifying the kind and intensity of plant disease by observing changes in specific plant parts, such as the color of the leaf, which is how the diagnosis is traditionally made. The use of visual assessments has been shown effective over time [9]. However, there is a significant amount of intricacy involved in diagnosing a plant through visual observation of its physical properties [10]. Due to their high level of intricacy, skilled plant pathologists and agronomists may misidentify the illnesses in plants.
As the first measures needed before an effective disease management strategy is developed, a prompt and accurate evaluation is required for the disease to be managed effectively. A poorly thought-out disease management strategy may arise from an incorrect diagnosis or a delayed diagnosis, both of which increase the risk of agricultural crop loss. Anybody, from inexperienced farmers to seasoned agronomists, would benefit from an efficient disease detection system when identifying afflicted plants based solely on visual inspection of the leaves [11].
Additionally, the ability for farmers to easily access an effective diagnostic system having the format of a mobile application could prove highly advantageous, since it can be particularly beneficial in regions of the globe where the availability of specialized tools is limited, such as screening techniques based on genome sequencing and microscopy, is limited due to financial and infrastructural challenges. The growing capacity of computer technologies, like Graphical Processing Unit (GPU) mechanisms, has made it possible for new theories and methodologies to be developed, leading to the phenomenal rise of machine learning systems in the previous few years. Among these are Artificial Neural Network (ANN) structures used in deep learning algorithms with many processing layers as opposed to typical neural network approaches with few processing layers [12]. Deep learning has garnered a lot of attention lately because it can do better than previous models in supervised and unsupervised pattern analysis and classification [13]. Deep learning models have been successfully used in many domains, such as image identity, mining data, as well as areas needing a high level of complexity [14].
In agriculture, deep learning structures leveraging Convolutional Neural Networks (CNNs) were applied in cases such as fruit counting, fruit detection, plant identification, and, most notably, disease diagnosis [15]. CNNs are among the most potent methods for modeling intricate processes and for pattern identification in applications such as visual pattern recognition that require massive amounts of data. This is because CNNs can identify patterns in the incoming images without requiring feature engineering of any kind, which spares the images from needless and involved preprocessing [16]. The authors in [17] reported that over 60% of developed countries have machine-learning applications. These applications are currently in use for cutting-edge tasks like diagnosing diabetes [18], portable speech recognition, and conversational systems, like Alexa from Amazon, Siri from Apple, Cortana from Microsoft, and Google Assistant. Considering that many farmers in underdeveloped countries do not have access to the latest innovations needed for the prompt and precise diagnosis needed to facilitate the timely detection of plant diseases, its use for plant disease diagnostics should be encouraged. Additionally, due to the recent global proliferation of mobile phones, farmers in underdeveloped nations now have access to cell phones that may one day be used by them as a diagnostic tool [19].
Early identification of plant diseases in the initial stages of small green leaf growth is essential. The financial stability of farmers is closely tied to the production of high-quality crops, which relies on the healthy development of these plants in the agricultural field. These days, early-stage disease identification is an essential component of research. Early on in their development, little green-leaf plants are particularly vulnerable to disease. Identifying plant diseases is among the most fundamental agricultural tasks. The majority of the time, identification is done by hand, visually, through molecular and serological testing, or microscopy. Early-stage plant disease detection techniques have proven to be more beneficial and productive in the context of agriculture. Green plant sickness essentially develops quickly across the leaves, stems, and branches of little green plants, stunting their growth and making it impossible for any manual method to identify the illness at an early stage. Therefore, certain automatic disease detection computational approaches should be developed to identify and detect early-stage diseases utilizing the different images of plant leaves to facilitate the timely identification of green plant disease. Thus, this study concentrates on the implementation of a web-based tomato plant disease detection and recognition system using transfer learning leveraging CNN (DenseNet121). The proposed system has a Graphical User Interface (GUI) to identify tomato plant disease using images of a leaf, which allows users to upload images and also view results.
Related works
Farming has a vital role in the global economy as a key means of food, revenue, and jobs. Nigeria's GDP, one of the developing nations in Africa, is primarily derived from agriculture, which employs 37.99% of the workforce in 2022 and contributed 24.45% in 2016 and 25.70% in 2020 [20] to the country's GDP. This is similar to various other developing nations with large populations of farmers. Since agriculture contributes significantly to national economic growth, plant diseases, and pest infections may harm the industry by lowering the quality of food produced. Treatments intended as preventive measures do not work to stop epidemics or endemic diseases. Crop quality losses could be avoided with timely recognition and accurate diagnosis of diseases through the use of an appropriate crop protection system. Plant disease identification is regarded as a critical issue and is very significant. Making better decisions while managing agricultural production may be achieved through early diagnosis of plant diseases.
Domesticating main food crops and animals began several decades back with the creation of agriculture. Food insecurity is a significant worldwide issue that humanity is currently facing [21], with plant diseases being a major contributing factor [22]. A global crop output loss of approximately 16% is estimated to be caused by plant diseases [23]. Around 50% of wheat and 26-29% of soybeans could be lost globally due to pests, according to estimates [23]. Plant infections can be classified into several broad categories, including bacteria, viruses, nematodes, algae, viroid infections, fungus-like microbes, and parasitic plants [24]. Examples of areas where Artificial Intelligence (AI), Machine Learning (ML), and computer vision have been very helpful are electricity prediction from renewable resources and medical devices. When the COVID-19 epidemic struck, Artificial Intelligence (AI) was significantly utilized in prognostic applications and the detection of lung-related disorders [25]. Early identification and evaluation of plant illnesses allow for similar cutting-edge technology to be utilized for its harmful impact reduction. Given how labor-intensive, time-consuming, and tiresome manual plant disease monitoring is, there has been a lot of research done recently on using computer vision and artificial intelligence to automatically recognize and evaluate infections in plants. Plant disease could be automatically identified and classified with an accuracy of 83.07% using a Bacterial Foraging Optimization-based Radial Basis Function Network (BRBFNN), according to Sidharth, et al. a wide range of tasks involving computer vision in several fields have successfully employed the Convolutional Neural Network (CNN), a widely common neural network architecture [12]. Plant disease categorization and detection have been studied using CNN architecture and its variations.
In their comparison of several CNN architectures, Sunayana, et al. [26] were successful in identifying infection in mango and potato leaves, achieving 98.33% accuracy with AlexNet and 90.85% accuracy with a shallow CNN model. Using a pre-trained VGG16 model, Guan, et al. [27] estimated the disease severity in apple plants with an accuracy of 90.40%. Chowdhury, et al. [24] used the LeNet model to classify healthy and diseased banana leaves with a 99.72% accuracy rate. Tomatoes are one of the most significant food crops in the world, consuming 20 kg per person per year and making up 15% of all vegetable intake on average. Panchal, et al. [28] introduced a CNN-based deep learning model that can accurately classify plant illnesses. Model training used a dataset of 87,000 RGB images that is accessible to the public. First, preprocessing and then segmentation was done. CNN was utilized to classify the data. 93.5% identification accuracy was achieved by this model; however, in later phases, the categories became confusing because it was unable to classify some of them. Furthermore, the limited accessibility of the data resulted in a decrease in the model's performance.
To increase detection accuracy, a hybrid convolutional neural network was proposed by [29] for categorizing plant diseases affecting bananas. Without altering any default settings, their preprocessing procedure for an initial raw image involved the employment of a median filter to maintain the traditional image dimensions. Using this strategy, both CNN and a fusion SVM were used. Using a multiclass Support Vector Machine (SVM), it was possible to identify the type of disease or infection in banana leaves that were affected in Phase 2 after it had been used in Phase 1 to classify the leaves as either healthy or infected. Following categorization, the support vector machine was fed the input and it produced 99% accurate classifications. CNN produced data that were more accurate than those using traditional methods, but it lacked diversity, according to earlier studies. Jadhav, et al. [30] introduced a CNN for diagnosing plant diseases. This strategy used CNN models that had already been trained to find illnesses in soybean plants. Although pre-trained transfer learning techniques like GoogleNet, AlexNet, and GoogleNet yielded better results in the tests, the model lacked categorization diversity. Many of the models that are now in use concentrate on identifying specific plant illnesses rather than creating a model to categorize different plant problems. The main reason is that there aren't enough databases to train deep-learning models based on a variety of plant species.
Using images from a poor-quality sample as a starting point, Abayomi-Alli, et al. [31] were the first to suggest a revolutionary histogram modification strategy that improved the recognition accuracy of deep learning models. This study aimed to improve the photographs in the cassava leaf disease dataset by applying motion blurring, overexposure, resolution down-sampling, and Gaussian blurring using a modified MobileNetV2 neural network model. To circumvent the data scarcity that a data-hungry deep-learning network faces during the training period, they produced synthetic pictures with modified color value distributions. Using a technique akin to that of Abayomi-Alli, et al. [31], Abbas, et al. [32] created a database of simulated photos of tomato plant leaves using a conditional generative adversarial network. With the use of generative networks, real-time data gathering and acquisition—which were previously costly, challenging, and time-consuming—is now feasible. Anh, et al. [33] suggested a multi-leaf system of classification using a previously trained MobileNet CNN model and a standard dataset. With a constant accuracy of 96.58%, the model did well in categorization. Furthermore, Kabir, et al. [34] built a multi-label CNN using transfer learning techniques such as DenseNet, MobileNet, Inception, VGG, ResNet, and Xception for the categorization of different plant ailments. This is the first study, according to the authors, that uses a multi-label CNN to classify 28 different plant disease groups. A study by Astani, et al. [35] suggested classifying plant infections by leveraging the Ensemble Classifier. Taiwan Tomato Leaf and Plant Village datasets were employed to examine the performance of the peak ensemble classifier.
Pradeep, et al. [36] suggested the EfficientNet model and used a convolutional neural network for multi-class and multi-label classification. The identification of plant diseases was more effectively influenced by the CNN covert network. When benchmark datasets were used for validation, the model did not perform as well. PlantVillage, a publicly available benchmark dataset, was utilized in [37] to propose an efficient, durable, loss-fused Convolutional Neural Network (CNN) that produced a 98.93% classification accuracy. Despite the classification accuracy being increased by this strategy, the real-time photos were used in different situations, and the model performed terribly. Later, a transfer learning strategy CNN network was presented by Enkvetchakul and Surinta [38] for two plant diseases. The best accurate prediction outcomes were obtained by using two pre-trained network models, namely MobileNetV2 and NASMobileNet, for the classification of plant diseases. Deep learning can address overfitting by utilizing the data augmentation technique. To apply the data augmentation methods, an experimental configuration featuring rotation, shift, brightness, cut-out, mix-up, and zoom was used. We used two different kinds of datasets: databases about leaf diseases and Cassava 2019. At 84.51%, the maximum test accuracy was obtained after the assessment.
Summary
From the literature reviewed, while much work has been done on classifying and identifying diseases in plant leaves, particularly in tomatoes and other plants, there hasn't been much focus on how reliable leaf images with different backgrounds are used for large picture classes. Photographs taken in real life might differ greatly from one another in terms of lighting, clarity of the image, alignment, and other factors. This study uses a trained model to address this issue.
By harnessing online technology, the designed web-based system seeks to increase agricultural output and efficiency by supporting farmers in making knowledgeable crop selection decisions. Furthermore, Convolutional Neural Networks (CNNs), in particular, are machine learning algorithms that are used for disease identification, ensuring accurate and timely detection. In addition to its disease recognition functionality, the web-based system provides a user-friendly interface accessible through web browsers. Farmers can upload pictures of the leaves on their tomato plants to identify diseases. After submission, the image is processed by the system using machine learning algorithms to assess and diagnose any diseases found, giving farmers comprehensive information about each ailment and suggestions for how to treat it. The web-based system streamlines the process of disease identification, empowering farmers to diagnose plant health issues quickly and accurately. By providing a seamless interface for image upload and disease analysis, the system enhances decision-making capabilities and supports proactive management of crop health.
The six steps involved in accurately and successfully diagnosing plant diseases were covered in this section.
Data collection
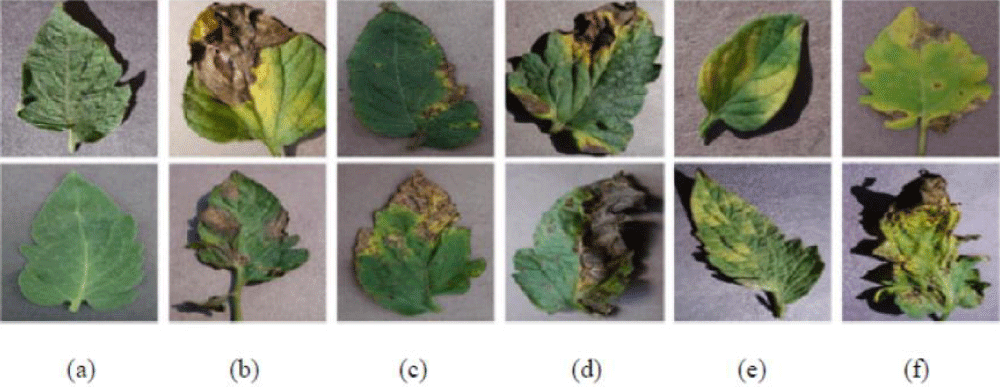
A dataset of 11165 images forming seven classes of tomato diseases (such as healthy, Leaf mold, yellow leaf curl virus, bacterial spot, late blight, early blight, and mosaic virus) was used for CNNs training. The images used were obtained from the PlantVillage dataset (https://www.kaggle.com/datasets/noulam/tomato). The images used in training are represented in Figure 1.
Figure 1: Plant disease images from the PlantVillage dataset. (a) Healthy. (b) Late blight. (c) Bacterial spot. (d) Early blight. (e) Leaf Mold. (f) Septoria leaf spot.
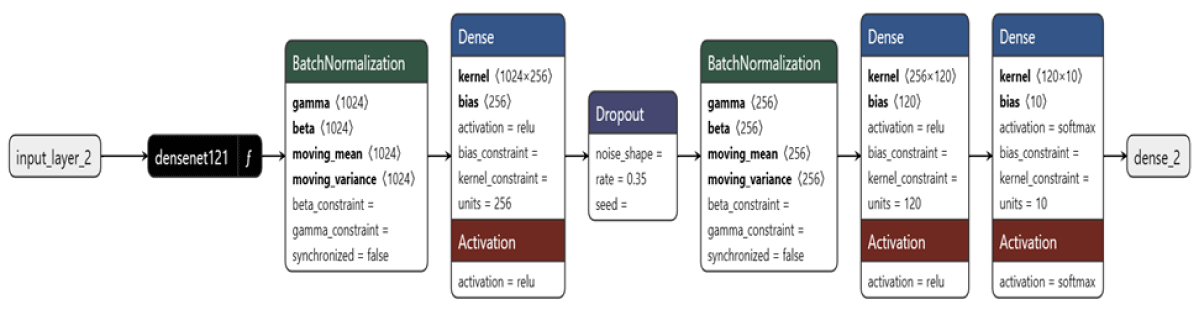
Figure 2: Model Architecture for the DenseNet121 Model.
Preprocessing
Identification of plant diseases is thought to start with image preprocessing. Numerous preprocessing procedures are involved, including segmenting disease regions, morphological operations, color modification, noise removal, and image scaling. The Gaussian filter, the median filter, and the Wiener filter are just a few methods for eliminating noise. Some color models, including RGB, YCbCr, HSV, and CIEL, have been used in image preprocessing.
Feature extraction
One of the fundamental steps in machine learning is feature extraction. Convolutional layers detect basic to complex features (edges, textures, shapes) of the raw images of tomato plants while pooling layers reduce dimensionality. Fully connected layers then combine and refine these features into a final vector, which the output layer uses to make the final prediction.
Data augmentation
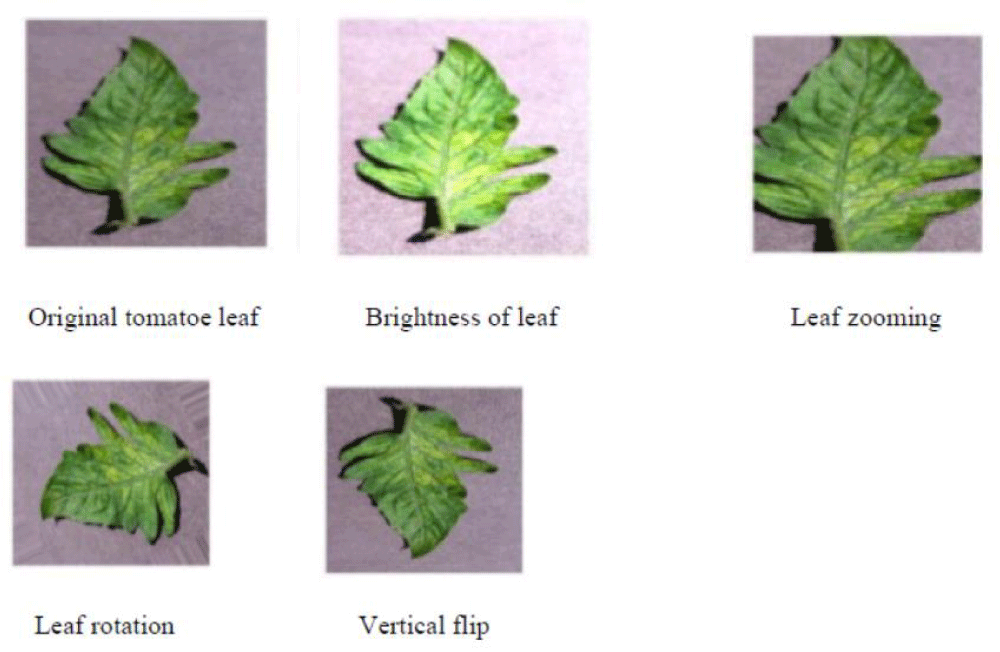
Data augmentation is a mathematical technique used to expand the dimensions of an image database. This process involves applying various mathematical operations, such as rotating the image by 90°, 180°, or 270°, translating (scaling), resizing the image, reducing its clarity, blurring it to different extents, altering its color and effects, performing geometric transformations, and flipping the image horizontally or vertically, among others (Figure 2).
Model training
This procedure guarantees a methodical assessment of the VGG16, VGG19, and DenseNet121 models through the use of model evaluation criteria including performance accuracy and loss value, ultimately assisting in the selection of the best model for real-world use. A comparison of the VGG16, VGG19, and DenseNet121 models is the result of this experimentation. The chosen DenseNet121 model for the given picture identification challenge was the one with the highest accuracy on the sample dataset.
The model architectural design
A model called DenseNet121 (Figure 3) was selected because of its excellent image categorization performance. It was selected due to its efficient architecture and high accuracy in image classification. Transfer learning was utilized by fine-tuning DenseNet121, which was pre-trained on ImageNet. The model was further trained on the tomato leaf dataset to adapt it to the particular classification assignment.
Figure 3: Process of Augmented Image.
Web application deployment
The image recognition model is trained and refined; it is deployed on the web to be accessible to users through a well-designed graphical interface. An online application with an intuitive interface was developed using this concept, allowing users to upload images of plants. The web application uses web technologies including HTML, CSS, and JavaScript to produce an intuitive layout that makes it easy for users to engage with the program. The backend, implemented using Django Framework, handles image uploads, passes them to the trained model, and returns the classification results. The graphical interface then displays the predicted tomato leaf disease along with a brief description. This deployment allows users to leverage the model's capabilities through a seamless and interactive web experience, making sophisticated image recognition accessible and practical.
Model performance evaluation
The following evaluation metrics were used in this study:
Accuracy: The fraction of accurately predicted scenarios to the total scenarios. It's useful for equilibrium datasets.
(1)
Precision: The fraction of accurately predicted positive scenarios to the sum of the predicted positives. It shows how many of the forecasted positive instances are positive.
(2)
Recall (Sensitivity): The fraction of accurately forecasted positive instances to all real positives. It measures the capability of the model to pick out all useful instances.
(3)
F1 Score: The harmonic average of precision as well as recall, providing a single standard that poises both concerns.
(4)
Area Under the Curve (AUC) - Receiver Operating Characteristic (ROC): evaluates how well the model can distinguish between various categories. Greater AUC denotes a superior model.
Entropy loss function: When a model assigns low probabilities to the proper class, it is penalised more heavily than when it assigns true labels. This function is often referred to as cross-entropy loss. It measures the difference between the true labels and the forecasted probabilities. To increase prediction accuracy, it is frequently applied to classification jobs.
Implementation
Experimental setting: A dataset of 11165 images forming seven classes of tomato was obtained from the PlantVillage dataset (https://www.kaggle.com/datasets/noulam/tomato).
Image preprocessing includes segmenting disease regions, morphological operations, color modification, noise removal, and image scaling. Preprocessing the images is necessary for deep learning techniques; this entails expanding them in size from 256 × 256 to 224 x 224 for the beforehand train models and the recommended method. This was a result of the previous taught models having been trained in dimensions of 224 × 224. Divide the photos by 255 to raster them to fit the network's initial values. Make 20% test and verification subsets and 80% training subsets out of the dataset. To train models using a set of field pictures that had more properties than laboratory images contained in the dataset, 500 produced photographs were added to the evaluation dataset, and 129 field photos were added to an assortment of field photographs acquired online. Using their expertise in artificial neural networks, the Gaussian Noise, Gaussian Blur, and Median filters are applied independently to the obtained dataset to distinguish clearly between their performances on the dataset and extract useful features. Thereafter, convolutional layers were used in the detection of basic to complex features (edges, textures, shapes) of the raw images of tomato plants, while pooling layers reduced dimensionality. Fully connected layers then combine and refine these features into a final vector, which the output layer uses to make the final prediction. To create photographs, we carried out an augmentation procedure by using metrics such as brightness with a value between (0.5 - 1) and rotation with 45° and 90°, as well as flipping horizontally and vertically and zooming with 0.5. The training dataset is expanding to include the 500 produced photos. A dataset's single image with augmented types is shown in Figure 2, a method for augmenting data to identify crop leaf illnesses. Using both the Binary Mask and the Pre-Activation Residual Attention Block (PaRAB), the plant data augmentation strategy proposed in this work aims to improve the expression capability of the model. This was investigated by taking inspiration from the Cycle GAN structure. These enhancements enable the suggested model to produce plant leaf data that is aesthetically pleasing. By generating insufficiently numerous affected samples from typical plant leaves, the suggested strategy increases the diversity of samples and addresses the bias issue with early diagnostic models for plants.
In the model training stage for image recognition, input images and their labels were fed into the model, which extracts features through its layers. The model then makes predictions that were evaluated against the real tags through a loss function to measure accuracy. When backpropagation occurs, the model modifies its weights according to the calculated loss to improve predictions. This iterative process is repeated over multiple epochs to enhance performance, while regular validation guarantees that the model performs properly when applied to new, untested data. Overall, this stage focuses on refining the model to accurately classify images by learning from and adapting to the provided dataset. After the model training, the best model was selected. The trained DenseNet121 model was saved in `.h5` format and loaded into the Django application using the `predict.py` script. The `predict.py` script is integral to the application as it loads the pre-trained DenseNet121 model from a saved `.h5` file and uses it to make predictions on the uploaded images. The integration between the model and the Django application is seamless
i. When a user uploads an image, it is processed and passed to the `predict.py` script.
ii. The script preprocesses the image to the required input format for the DenseNet121 model, performs the prediction, and returns the result.
iii. The result is then handled by `views.py`, which renders the appropriate template to display the result back to the user. The `views.py` file manages user requests and uses the model to make predictions on uploaded images.
Activity diagram
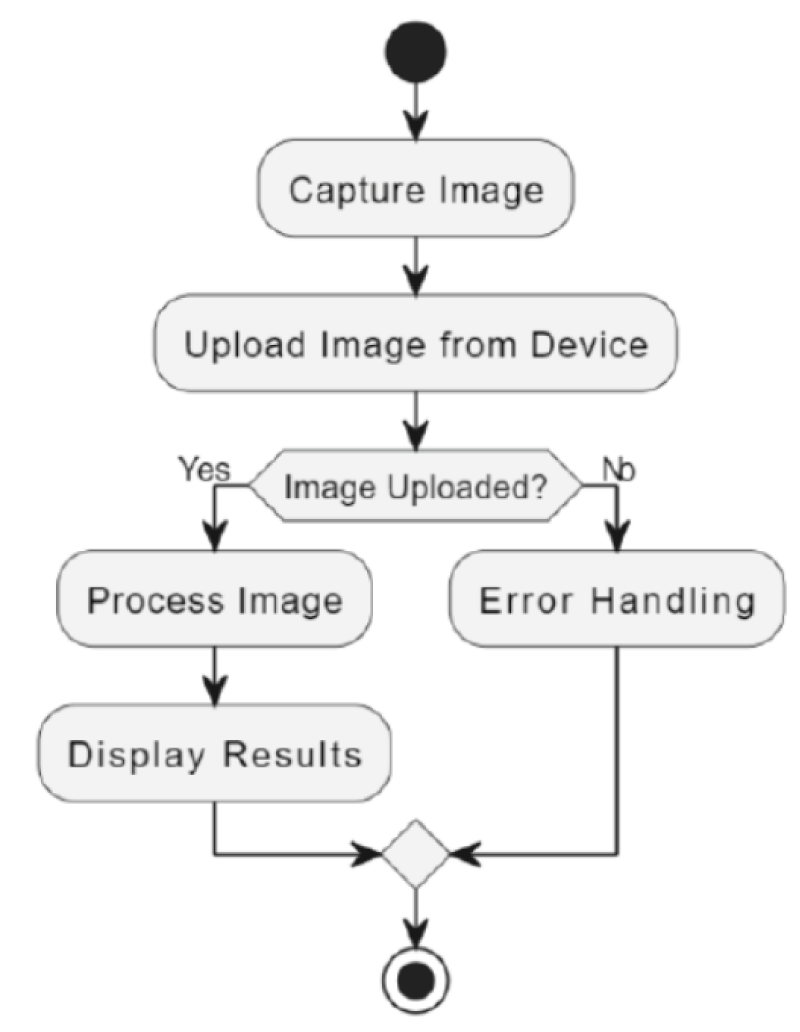
The activity diagram in Figure 4 illustrates the process of capturing or uploading an image and displaying the results. It begins when the user selects either "Capture Image" or "Upload Image." If "Capture Image" is chosen, the web application activates the camera, allowing the user to capture an image, which is then received by the web application. If "Upload Image" is selected, the web application opens a file selection dialog for the user to choose an image from their device, which is then uploaded. The web application processes the uploaded image, and finally, displays the results. The process concludes with the end of the activity.
Figure 4: Activity Diagram of the web-based Tomato Plant Disease Detection System.
The outcomes of a Django web application created to categorize tomato leaf diseases using a DenseNet121 model were reported in this section. Given its superior performance accuracy, the DenseNet121 model was selected.
Model comparison result of VGG16, VGG19 and DenseNet121: For comparison among the models, the performance Accuracy and Entropy loss function (Loss Value) were used to measure performance in addition to selecting the ideal model for the task. The result is displayed in Table 1.
| Table 1: Model Comparison Result of VGG16, VGG19, and DenseNet121. | ||||
| Model | Training Accuracy | Validation Accuracy | Training Loss | Validation Loss |
| DenseNet121 | 98.90 | 98.28 | 0.0332 | 0.0555 |
| VGG16 | 78.37 | 87.54 | 0.6020 | 0.3679 |
| VGG19 | 75.20 | 84.62 | 0.7456 | 0.4713 |
The DenseNet121 evaluation result
The trained model was further evaluated to determine its effectiveness in classifying tomato leaf diseases. The results are shown in Figures 5-7 and a summary of the results is presented in Table 2.
| Table 2: DenseNet121 Evaluation Result. | |
| Metric | Value |
| Accuracy | 98.28% |
| Loss | 0.0555 |
| AUC | 0.9990 |
| Precision | 0.9830 |
| Recall | 0.9817 |
| F1 Score | 0.9618 |
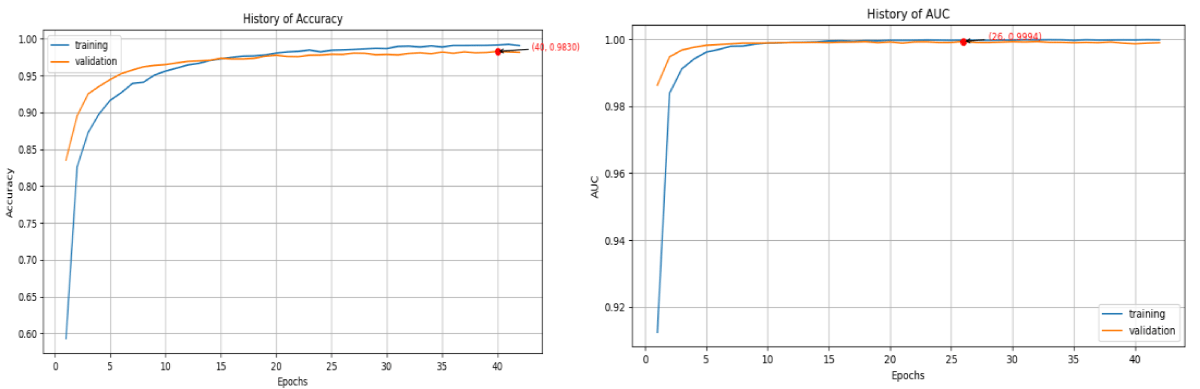
Figure 5. Graphical representation of Accuracy and AUC for DenseNet121. The accuracy curve is on the left-hand side while AUC is on the right-hand side. The orange color denotes the validation curve while the blue is the training curve.
Figure 5: Graphical representation of Accuracy and AUC for DenseNet121. The accuracy curve is on the left-hand side while AUC is on the right-hand side. The orange color denotes the validation curve while the blue is the training curve.
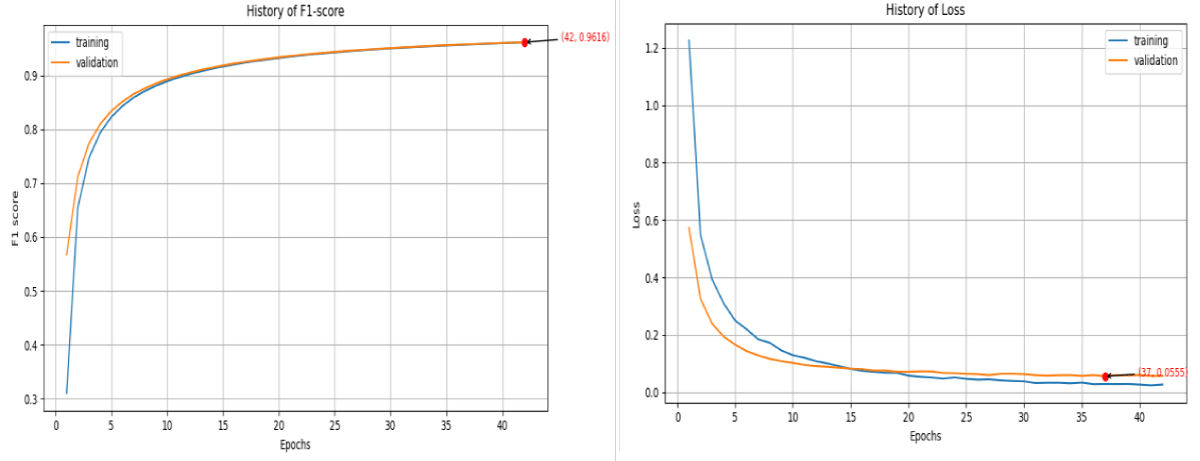
Figure 6. Graphical representation of F1-score and Loss value for DenseNet12. The F1 score curve is on the left-hand side while the loss function curve is on the right-hand side. The orange color denotes the validation curve while the blue is the training curve.
Figure 6: Graphical representation of F1-score and Loss value for DenseNet12. The F1 score curve is on the left-hand side while the loss function curve is on the right-hand side. The orange color denotes the validation curve while the blue is the training curve.
Figure 7. Graphical representation of precision for DenseNet121. The orange color denotes the validation curve while the blue is the training curve.
Figure 7: Graphical representation of precision for DenseNet121. The orange color denotes the validation curve while the blue is the training curve.
User interface
Web upload: The web form designed for image upload is straightforward and user-friendly. Users can select an image file from their device and submit it for analysis. The web app interface is shown in Figure 8.
Figure 8: Web-based Interface Image Upload.
Results display
After processing the uploaded image, the classification result is dynamically displayed on the web page, providing immediate feedback to the user about the health status of the tomato leaf as depicted in Figure 9.
Figure 9: Result Display of the detection and Recognition of Early Blight Disease in Tomato Plant leaf with an Accuracy of 87%.
The present study employed Transfer learning to build a web-based system for detecting and identifying tomato plant diseases. VGG16, VGG19, and DenseNet121 were the three models that were employed using the CNN architecture. In identical training and testing settings, these three models were chosen based on their best performance. Since it performed so well in image classification tasks, the DenseNet121 model was selected. Due to its excellent classification accuracy and effective architecture, it was chosen. This was evidenced in the result obtained as presented in Table 1 with higher scores in training and validation accuracy while it has minimum loss function as compared with the other two models. The loss function is about the high accuracy in predicted values as compared with actual values. With this result, DenseNet121 was most preferred for this system. Transfer learning was utilized by fine-tuning DenseNet121, which was pre-trained on ImageNet. The model was further trained on the tomato leaf dataset to adapt it to the particular classification assignment.
The DenseNet121 model was further evaluated after the implementation of the system. This model designed for disease detection demonstrates impressive performance metrics. It demonstrates a high degree of expertise in predicting the existence of the disease from images, as demonstrated by its 98% accuracy rate in Table 2. This high accuracy highlights the model's capacity to correctly identify the illness in the great majority of instances. Furthermore, the excellent precision of the model indicates a low false positive rate, implying that it hardly ever misidentifies healthy plants as sick. The model's ability to accurately identify all pertinent disease cases is also indicated by its high recall value ensuring that few, if any, diseased plants go undetected. The high F1 score, a harmonic mean of precision and recall, confirms that the model maintains a substantial balance between these two metrics, highlighting its robustness and reliability. As illustrated in Figure 5, the model's accuracy improves progressively with each epoch. This trend signifies effective learning, as the model becomes increasingly adept at recognizing patterns associated with the disease over time. The consistent increase in accuracy demonstrates that the model is successfully adapting and refining its predictive capabilities with more training.
Result comparison
Several studies use artificial intelligence for plant disease recognition and classification. Panchal, et al. [28] introduced a CNN-based deep learning model that can accurately classify apple plant illnesses, and Jadhav, et al. [30] introduced a CNN for diagnosing soybean plant diseases. This strategy used CNN models that had been trained to find illnesses in soybean plants. Narayanan, et al. used primary data from India to classify banana plant disease using CNN and a fused SVM-based classifier. Abass, et al. and Astani, et al. used PlantVillage datasets for tomato plant disease classification. It can be seen in Table 3 that different levels of accuracy were achieved with different models for plant disease recognition. This study has included a web-based application with a deep learning model to facilitate easy tomato plant disease detection by the farmers. The proposed system was tested with real-time tomato leaf disease images for recognition and diagnosis and the result was good. The summary of the result comparison of the existing works with our proposed system is presented in Table 3.
| Table 3: Comparison of the Proposed System with the Existing Works. | |||||
| Author | Dataset | Plant considered | Model | Accuracy | Real-time detection through GUI |
| Panchal, et al. [28] | PlantVillage | Apple | VGGNet | 93.5% | N/A |
| Jadhav, et al. [30] | Kaggle | soybean leaves | AlexNet and GoogleNet CNNs | AlexNet: 98.75% | N/A |
| Narayanan et al. [29] | Primary data from South India | Banana | CNN + fusion SVM-based classifier | 99% | N/A |
| Abbas, et al. [32] | PlantVillage and synthetic data | Tomatoes | ResNet, DenseNet121 | 97% | N/A |
| Astani, et al. [35] | Plantvillage and Taiwan tomato leaves | Tomatoes | Ensemble learning | 96% | N/A |
| Proposed model | PlantVillage and local Nigerian tomato leaves | Tomatoes | VGG16, VGG19, and DenseNet121 | 98% | Web-based system |
This study focused on developing a web-based plant disease detection and recognition system using machine learning techniques, specifically targeting diseases affecting tomato plants. Plant afflictions give way to a substantial problem in agricultural productivity and the livelihood of farmers. Punctual and accurate diagnosis is crucial for effective disease management and can significantly reduce crop loss and associated economic impacts. The research work utilized a deep learning method, particularly Convolutional Neural Networks (CNNs), for the recognition of plant diseases. By leveraging transfer learning techniques, tomato crop photographs were used for model training to recognize various diseases. The implementation included the implementation of a Graphical User Interface (GUI) that allows users to upload images of tomato leaves, which the system then analyses to detect any disease presence. The system was designed to be accessible and user-friendly, providing farmers in regions with limited access to advanced diagnostic tools, with a reliable means to identify plant diseases early. The model achieved 98% accuracy which is impressive. The work has improved decision-making in agricultural practices, thereby enhancing productivity and reducing losses due to plant diseases. This work has supported the sustainable development goal (SDG 1), “zero to hunger”.
Recommendation
For farmers, especially those in developing regions, the following recommendations can enhance the utility of the developed system:
i. Regular monitoring: Farmers should regularly monitor their crops and use the detection system to identify any signs of disease early. This proactive approach can help in taking timely action to manage and control the spread of diseases.
ii. Training and education: It is crucial to teach farmers how to operate the system correctly. This entails being aware of how to take clear images of the plant leaves and comprehend the meaning behind the system's output.
iii. Integration with traditional methods: While the system provides a modern approach to disease detection, it could be utilized for complete disease management in connection with conventional techniques and professional guidance.
iv. Feedback loop: It would be beneficial for farmers to offer input on how the system is working so that the model's accuracy may be continuously enhanced.
v. Access to resources: Ensuring farmers have access to the necessary resources, such as smartphones and internet connectivity, is crucial for the widespread adoption of the system.
Authors’ contribution
T. E. has the research idea and design, A. M. reviewed the literature, and the implementation was done by M. J. and E. J. The manuscript was written by E. J. and the review was done by T. E.
- Mngoma MF, Magwaza LS, Sithole NJ, Magwaza ST, Mditshwa A, Tesfay SZ,et al. Effects of stem training on the physiology, growth, and yield responses of indeterminate tomato (Solanum lycopersicum) plants grown in protected cultivation. Heliyon. 2022;8(5). Available from: https://doi.org/10.1016/j.heliyon.2022.e09343
- Moran NE, Thomas-Ahner JM, Wan L, Zuniga KE, Clinton SK. Tomatoes, lycopene, and prostate cancer: What have we learned from experimental models? J Nutr. 2022;152(6):1381-1403. Available from: https://doi.org/10.1093%2Fjn%2Fnxac066
- Salau SA, Salman M. Economic analysis of tomato marketing in Ilorin metropolis, Kwara State, Nigeria. J Agric Sci. 2017;62(2):179-191. Available from: http://dx.doi.org/10.2298/JAS1702179S
- Agrios GN. Introduction. In: Agrios GN, editor. Plant Pathology. 5th ed. Elsevier; 2005;3-75. Available from: https://shop.elsevier.com/books/plant-pathology/agrios/978-0-08-047378-9
- Sibiya M, Sumbwanyambe M. A computational procedure for the recognition and classification of maize leaf diseases out of healthy leaves using convolutional neural networks. AgriEngineering. 2019;1(1):119-131. Available from: https://doi.org/10.3390/agriengineering1010009
- O’Brien PA. Biological control of plant diseases. Australas Plant Pathol. 2017;46(4):293-304. Available from: https://doi.org/10.1007/s13313-017-0481-4
- Lugtenberg B. Introduction to plant-microbe interactions. In: Lugtenberg B, editor. Principles of Plant-Microbe Interactions. Cham: Springer. 2015. Available from: https://link.springer.com/book/10.1007/978-3-319-08575-3
- Chakraborty S, Tiedemann A, Teng P. Climate change: Potential impact on plant diseases. Environ Pollut. 2000;108(3):317-326. Available from: https://doi.org/10.1016/s0269-7491(99)00210-9
- Bock CH, Chiang KS, Del Ponte EM. Plant disease severity estimated visually: A century of research, best practices, and opportunities for improving methods and practices to maximize accuracy. Trop Plant Pathol. 2022;47(1):25-42. Available from: http://dx.doi.org/10.1007/s40858-021-00439-z
- Ferentinos KP. Deep learning models for plant disease detection and diagnosis. Comput Electron Agric. 2018;145:311-318. Available from: http://dx.doi.org/10.1016/j.compag.2018.01.009
- Mohanty SP, Hughes DP, Salathé M. Using deep learning for image-based plant disease detection. Front Plant Sci. 2016;7:215. Available from: https://doi.org/10.3389/fpls.2016.01419
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436-44. Available from: https://doi.org/10.1038/nature14539
- Lawson CE, Martí JM, Radivojevic T, Jonnalagadda SVR, Gentz R, Hillson NJ, et al. Machine learning for metabolic engineering: A review. Metabol Eng. 2020;63:34-60. Available from: https://doi.org/10.1128/msystems.00925-21
- Nguyen G, Dlugolinsky S, Bobák M, Tran V, García ÁL, et al. Machine learning and deep learning frameworks and libraries for large-scale data mining: A survey. Artif Intell Rev. 2019;52(1):77-124. Available from: https://link.springer.com/article/10.1007/s10462-018-09679-z
- Xiao F, Wang H, Xu Y, Zhang R. Fruit detection and recognition based on deep learning for automatic harvesting: An overview and review. Agronomy. 2023;13(6):16-25. Available from: https://doi.org/10.3390/agronomy13061625
- Rayed ME, Islam SS, Niha SI, Jim JR, Kabir MM, Mridha M. Deep learning for medical image segmentation: State-of-the-art advancements and challenges. Informatics Med Unlocked. 2024;47:101504. Available from: https://doi.org/10.1016/j.imu.2024.101504
- Lee I, Shin YJ. Machine learning for enterprises: Applications, algorithm selection, and challenges. Bus Horiz. 2020;63(2):157-70. Available from: https://ideas.repec.org/a/eee/bushor/v63y2020i2p157-170.html
- Karan O, Bayraktar C, Gümüşkaya H, Karlık B. Diagnosing diabetes using neural networks on small mobile devices. Expert Syst Appl. 2011;39(1):54-60. Available from: http://dx.doi.org/10.1016/j.eswa.2011.06.046
- Domingues T, Brandão T, Ferreira JC. Machine learning for detection and prediction of crop diseases and pests: A comprehensive survey. Agriculture. 2022;12(9):13-50. Available from: https://doi.org/10.3390/agriculture12091350
- Micheal VA. Impact of agricultural variables on gross domestic product (GDP) in Nigeria: A SARIMA approach. Afr J Agric Sci Food Res. 2024;14(1):78-91. Available from: http://dx.doi.org/10.62154/r567wk89
- Wells JCK, Stock JT. Life history transitions at the origins of agriculture: A model for understanding how niche construction impacts human growth, demography, and health. Front Endocrinol. 2020;11:325. Available from: https://doi.org/10.3389/fendo.2020.00325
- Strange RN, Scott PR. Plant disease: A threat to global food security. Annu Rev Phytopathol. 2005;43:83-116. Available from: https://doi.org/10.1146/annurev.phyto.43.113004.133839
- Oerke EC. Losses to pests. J Agric Sci. 2006;144(1):31-43. Available from: http://dx.doi.org/10.1017/S0021859605005708
- Chowdhury ME, Rahman T, Khandakar A, Ayari MA, Khan AU, Khan MS, et al. Automatic and reliable leaf disease detection using deep learning techniques. AgriEngineering. 2021;3(2):294-312. Available from: https://www.mdpi.com/2624-7402/3/2/20#
- Tzeng S, Hsieh C, Su L, Hsieh H, Chang C. Artificial intelligence-assisted chest X-ray for the diagnosis of COVID-19: A systematic review and meta-analysis. Diagnostics. 2023;13(4):1-18. Available from: https://doi.org/10.3390/diagnostics13040584
- Arya S, Sing R. A comparative study of CNN and AlexNet for detection of disease in potato and mango leaf. In: International Conference on Issues and Challenges in Intelligent Computing Techniques (ICICT). Ghaziabad, India. 2019. Available from: https://doi.org/10.1109/ICICT46931.2019.8977648
- Wang G, Sun Y, Wang J. Automatic image-based plant disease severity estimation using deep learning. Comput Intell Neurosci. 2017;2017:1-8. Available from: https://doi.org/10.1155/2017/2917536
- Panchal AV, Patel SC, Bagyalakshmi K, Kumar P, Khan IR, Soni M. Image-based plant diseases detection using deep learning. Mater Today Proc. 2022. Available from: http://dx.doi.org/10.1016/j.matpr.2021.07.281
- Narayanan KL, Krishnan RS, Robinson YH, Julie EG, Vimal S, Saravanan V, Kaliappan M. Banana plant disease classification using hybrid convolutional neural network. Comput Intell Neurosci. 2022;2022:1-22. Available from: http://dx.doi.org/10.1155/2022/9153699
- Jadhav SB, Udupi VR, Patil SB. Identification of plant diseases using convolutional neural networks. Int J Inf Technol. 2021;13:2461-2470. Available from: http://dx.doi.org/10.1007/s41870-020-00437-5
- Abayomi-Alli OO, Damaševičius R, Misra S, Maskeliūnas R. Cassava disease recognition from low-quality images using enhanced data augmentation model and deep learning. Expert Syst. 2021;38(4). Available from: http://dx.doi.org/10.1111/exsy.12746
- Abbas A, Jain S, Gour M, Vankudothu S. Tomato plant disease detection using transfer learning with C-GAN synthetic images. Comput Electron Agric. 2021;187:106279. Available from: http://dx.doi.org/10.1016/j.compag.2021.106279
- Eunice J, Popescu DE, Chowdary MK, Hemanth J. Deep learning-based leaf disease detection in crops using images for agricultural applications. Agronomy. 2022;12(10):2395. Available from: https://doi.org/10.3390/agronomy12102395
- Kabir MM, Ohi AQ, Mridha MF. A multi-plant disease diagnosis method using convolutional neural network. In: Computer Vision and Machine Learning in Agriculture: Algorithms for Intelligent Systems. Singapore: Springer; 2021. Available from: https://doi.org/10.48550/arXiv.2011.05151
- Astani M, Hasheminejad M, Vaghefi M. A diverse ensemble classifier for tomato disease recognition. Comput Electron Agric. 2022;198:107054. Available from: https://doi.org/10.1016/j.compag.2022.107054
- Prodeep AR, Morshedul Hoque ASM, Kbir MM, Rahman MS, Mridha MF. Plant disease identification from leaf images using deep CNN’s EfficientNet. In: International Conference on Decision Aid Sciences and Applications (DASA); 2022; Chiangrai, Thailand. Available from: http://dx.doi.org/10.1109/DASA54658.2022.9765063
- Gokulnath B, Usha Devi G. Identifying and classifying plant disease using resilient LF-CNN. Ecol Informatics. 2021;63:101283. Available from: http://dx.doi.org/10.1016/j.ecoinf.2021.101283
- Enkvetchakul P, Surinta O. Effective data augmentation and training techniques for improving deep learning in plant leaf disease recognition. Appl Sci Eng Prog. 2022;15(3):1-12. Available from: http://dx.doi.org/10.14416/j.asep.2021.01.003